#> $names
#> [1] "coefficients" "residuals" "effects" "rank"
#> [5] "fitted.values" "assign" "qr" "df.residual"
#> [9] "xlevels" "call" "terms" "model"
#>
#> $class
#> [1] "lm"Deep Learning
Neural Networks
Neural Networks
Stochastic Gradient Descent
R6
TensorFlow
Torch
R Code
Single Layer R Code
Multilayer R Code
Neural Networks
Neural networks are a type of machine learning algorithm that are designed to mimic the function of the human brain. They consist of interconnected nodes or “neurons” that process information and generate outputs based on the inputs they receive.
Uses
Neural networks are typically used for tasks such as image recognition, natural language processing, and prediction. They are capable of learning from data and improving their performance over time, which makes them well-suited for complex and dynamic problems.
Single Layer Neural Networks
A single layer neural networks can be formulated as linear function:
\[ f(X) = \beta_0 + \sum^K_{k=1}\beta_kh_k(X) \]
Where \(X\) is a vector of inputs of length \(p\) and \(K\) is the number of activations, \(\beta_j\) are the regression coefficients and
\[ h_k(X) = A_k = g(w_{k0} + \sum^p_{l1}w_{kl}X_{l}) \]
with \(g(\cdot)\) being a nonlinear function and \(w_{kl}\) are the weights.
Nonlinear (Activations) Function \(g(\cdot)\)
Sigmoidal: \(g(z) = \frac{e^z}{1+e^z}\)
ReLU (rectified linear unit): \(g(z) = (z)_+ = zI(z\geq0)\)
Single Layer Neural Network

Multilayer Neural Network
Multilayer Neural Networks create multiple hidden layers where each layer feeds into each other which will create a final outcome.
Multilayer Neural Network
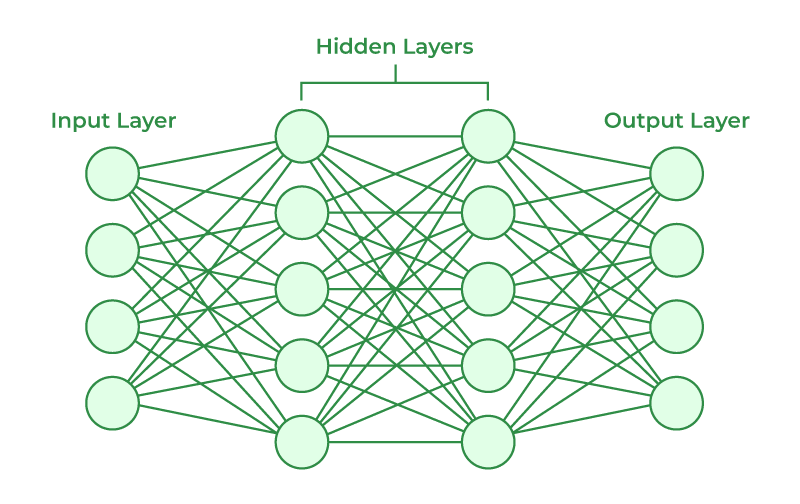
Hidden Layer 1
With \(p\) predictors of \(X\):
\[ h^{(1)}_k(X) = A^{(1)}_k = g\left\{w^{(1)}_{k0} + \sum^p_{j=1}w^{(1)}_{kj}X_{j}\right\} \] for \(k = 1, \cdots, K\) nodes.
Hidden Layer 2
\[ h^{(2)}_l(X) = A^{(2)}_l = g\left\{w^{(2)}_{l0} + \sum^K_{k=1}w^{(2)}_{lk}A^{(1)}_{k}\right\} \] for \(l = 1, \cdots, L\) nodes.
Hidden Layer 3 +
\[ h^{(3)}_m(X) = A^{(3)}_l = g\left\{w^{(3)}_{m0} + \sum^L_{l=1}w^{(3)}_{ml}A^{(2)}_{l}\right\} \] for \(m = 1, \cdots, M\) nodes.
Output Layer
\[ f_t(X) = \beta_{t0} + \sum^M_{m=1}\beta_{tm}h^{(3)}_m(X) \] for outcomes \(t = 1, \cdots, T\)
Stochastic Gradient Descent
Neural Networks
Stochastic Gradient Descent
R6
TensorFlow
Torch
R Code
Single Layer R Code
Multilayer R Code
Gradient Descent
This is an optimization algorithm used to identify the maximum or minimum of a functions.
Method
For a given \(F(\boldsymbol X)\), the minimum \(\boldsymbol X^\prime\) can be found by iterating the following formula:
\[ \boldsymbol X_{j+1} = \boldsymbol X_{j} - \gamma \nabla F(\boldsymbol X_j) \] where
\[ \boldsymbol X_j \rightarrow \boldsymbol X^\prime \]
- \(\nabla F\): gradient
- \(\gamma \in (0,1]\): is the step size
Stochastic Gradient Descent
Let:
\[ F(\boldsymbol X) = \sum^n_{i=1} f(y_i; \boldsymbol X) \]
Instead of minimizing \(F(\boldsymbol X)\) at each step, we minimize:
\[ F_\alpha(\boldsymbol X) = \sum_{i\in\alpha} f(y_i; \boldsymbol X) \] where \(\alpha\) indicates the data points randomly sampled to be used to compute the gradient.
R6
Neural Networks
Stochastic Gradient Descent
R6
TensorFlow
Torch
R Code
Single Layer R Code
Multilayer R Code
Object-Oriented Programming
Loosely speaking Object-Oriented Programming involves developing R objects with specialized attributes.
There are 3 main types of OOP: S3, R6, S4
#> # A tibble: 37 × 4
#> generic class visible source
#> <chr> <chr> <lgl> <chr>
#> 1 summary aov TRUE stats
#> 2 summary aovlist FALSE registered S3method
#> 3 summary aspell FALSE registered S3method
#> 4 summary check_packages_in_dir FALSE registered S3method
#> 5 summary connection TRUE base
#> 6 summary data.frame TRUE base
#> 7 summary Date TRUE base
#> 8 summary default TRUE base
#> 9 summary ecdf FALSE registered S3method
#> 10 summary factor TRUE base
#> # ℹ 27 more rowsR6
R6 is slightly different, where the generic functions belong to the object instead.
This allows us to modify an R objects with different methodologies if needed.
This is also useful when interacting with objects outside of R’s environment.
self
self is a special container within R6 that contains all the items and functions within the object.
You must always use self when dealing with R6 objects.
TensorFlow
Neural Networks
Stochastic Gradient Descent
R6
TensorFlow
Torch
R Code
Single Layer R Code
Multilayer R Code
TensorFlow
Tensorflow is an open-source machine learning platform developed by Google. Tensorflow is capable of completing the following tasks:
Image Classification
Text Classification
Regression
Time-Series
Keras
Keras is the API that will talk to Tensorflow via different platforms.
More Information
Torch
Neural Networks
Stochastic Gradient Descent
R6
TensorFlow
Torch
R Code
Single Layer R Code
Multilayer R Code
Torch
Torch is a scientific computing framework designed to support machine learning in CPU/GPU computing. Torch is capable of computing:
Matrix Operations
Linear Algebra
Neural Networks
Numerical Optimization
and so much more!
Torch
Torch can be accessed in both:
Pytorch
R Torch
R Torch
R Torch is capable of handling:
Image Recognition
Tabular Data
Time Series Forecasting
Audio Processing
More Information
R Code
Neural Networks
Stochastic Gradient Descent
R6
TensorFlow
Torch
R Code
Single Layer R Code
Multilayer R Code
Installation of Torch
Torch Packages in R
ISLR Torch Lab
ISLR uses Tensorflow.
Use this instead: https://hastie.su.domains/ISLR2/Labs/Rmarkdown_Notebooks/Ch10-deeplearning-lab-torch.html
Single Layer R Code
Neural Networks
Stochastic Gradient Descent
R6
TensorFlow
Torch
R Code
Single Layer R Code
Multilayer R Code
Penguin Data
Build a single-layer neural network that will predict body_mass_g with the remaining predictors except for year. The hidden layer will contain 50 nodes, and the activation functions will be ReLU.
Model Description
Creates the functions needed to describe the details of each network.
Optimizer Set Up
Fit a Model
Testing Model
Multilayer R Code
Neural Networks
Stochastic Gradient Descent
R6
TensorFlow
Torch
R Code
Single Layer R Code
Multilayer R Code
Penguins Data
Use the penguins data set to construct a 2-layer neural network to predict species with the other predictors, except for year. The 1st layer should have 10 nodes, the second layer should have 5 nodes, and use ReLU activations.
Model
modelnn2 <- nn_module(
initialize = function(input_size) {
self$hidden1 <- nn_linear(in_features = input_size,
out_features = 10)
self$hidden2 <- nn_linear(in_features = 10,
out_features = 5)
self$output <- nn_linear(in_features = 5,
out_features = 3)
self$drop1 <- nn_dropout(p = 0.4)
self$drop2 <- nn_dropout(p = 0.3)
self$activation <- nn_relu()
},
forward = function(x) {
x |>
self$hidden1() |>
self$activation() |>
self$drop1() |>
self$hidden2() |>
self$activation() |>
self$drop2() |>
self$output()
}
) initialize = function(input_size) {
self$hidden1 <- nn_linear(in_features = input_size,
out_features = 10)
self$hidden2 <- nn_linear(in_features = 10,
out_features = 5)
self$output <- nn_linear(in_features = 5,
out_features = 3)
self$drop1 <- nn_dropout(p = 0.4)
self$drop2 <- nn_dropout(p = 0.3)
self$activation <- nn_relu()
}Setup
Model Fitting
Testing
res <- fitted2 |>
predict(Xtesting) |>
torch_argmax(dim = 2) |>
as_array()
mean(as.numeric(Ytesting)==res)#> [1] 1



